elektronn2.neuromancer.loss module¶
-
class
elektronn2.neuromancer.loss.GaussianNLL(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeSimilar to squared loss but “modulated” in scale by the variance.
Parameters: Computes element-wise:
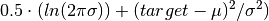
-
class
elektronn2.neuromancer.loss.BinaryNLL(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeBinary NLL node. Identical to cross entropy.
Parameters: Computes element-wise:
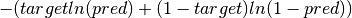
-
class
elektronn2.neuromancer.loss.AggregateLoss(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeThis node is used to average the individual losses over a batch (and possibly, spatial/temporal dimensions). Several losses can be mixed for multi-target training.
Parameters: - parent_nodes (list/tuple of graph or single node) – each component is some (possibly element-wise) loss array
- mixing_weights (list/None) – Weights for the individual costs. If none, then all are weighted
equally. If mixing weights are used, they can be changed during
training by manipulating the attribute
params['mixing_weights']. - name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
- The following is all wrong, mixing_weights are directly used (#) –
- losses are first summed per component, and then the component sums (The) –
- summed using the relative weights. The resulting scalar is finally (are) –
- such that (normalised) –
- The cost does not grow with the number of mixed components
- Components which consist of more individual losses have more weight e.g. If there is a constraint on some hidden representation with 20 features and a constraint the reconstruction of 100 features, the reconstruction constraint has 5x more impact on the overall loss than the constraint on the hidden state (provided those two loss are initially on the same scale). If they are intended to have equal impact, the weights should be used to upscale the constraint against the reconstruction.
-
class
elektronn2.neuromancer.loss.SquaredLoss(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeSquared loss node.
Parameters: - pred (Node) – Prediction node.
- target (T.Tensor) – corresponds to a vector that gives the correct label for each example. Labels < 0 are ignored (e.g. can be used for label propagation).
- margin (float or None) –
- scale_correction (float or None) – Downweights absolute deviations for large target scale. The value specifies the target value at which the square deviation has half weight compared to target=0 If the target is twice as large as this value the downweight is 1/3 and so on. Note: the smaller this value the stronger the effect. No effect would be +inf
- name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
-
class
elektronn2.neuromancer.loss.AbsLoss(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.loss.SquaredLossAbsLoss node.
Parameters: - pred (Node) – Prediction node.
- target (T.Tensor) – corresponds to a vector that gives the correct label for each example. Labels < 0 are ignored (e.g. can be used for label propagation).
- margin (float or None) –
- scale_correction (float or None) – Boosts loss for large target values: if target=1 the error is multiplied by this value (and linearly for other targets)
- name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
-
class
elektronn2.neuromancer.loss.Softmax(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeSoftmax node.
Parameters: - parent (Node) – Input node.
- n_class –
- n_indep –
- name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
-
class
elektronn2.neuromancer.loss.MultinoulliNLL(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeReturns the symbolic mean and instance-wise negative log-likelihood of the prediction of this model under a given target distribution.
Parameters: - pred (Node) – Prediction node.
- target (T.Tensor) – corresponds to a vector that gives the correct label for each example. Labels < 0 are ignored (e.g. can be used for label propagation).
- target_is_sparse (bool) – If the target is sparse.
- class_weights (T.Tensor) – weight vector of float32 of length
n_lab. Values:1.0(default),w < 1.0(less important),w > 1.0(more important class). - example_weights (T.Tensor) – weight vector of float32 of shape
(bs, z, x, y)that can give the individual examples (i.e. labels for output pixels) different weights. Values:1.0(default),w < 1.0(less important),w > 1.0(more important example). Note: if this is not normalised/bounded it may result in a effectively modified learning rate! - weakness (float) – Should be a number between 0 and 1. 0 (default) disables weak learning. If > 0, mix real targets with network outputs to achieve less accurate but softer training labels. Can improve convergence.
- following refers to lazy labels, the masks are always on a per patch basis, depending on the (The) –
- cube of the patch. The masks are properties of the individual image cubes and must be loaded (origin) –
- CNNData. (into) –
- mask_class_labeled (T.Tensor) – shape = (batchsize, num_classes).
Binary masks indicating whether a class is properly labeled in
y. If a classkis (in general) present in the image patches andmask_class_labeled[k]==1, then the labels must obeyy==kfor all pixels where the class is present. If a classkis present in the image, but was not labeled (-> cheaper labels), setmask_class_labeled[k]=0. Then all pixels for which they==kwill be ignored. Alternative: sety=-1to ignore those pixels. Limit case:mask_class_labeled[:]==1will result in the ordinary NLL. - mask_class_not_present (T.Tensor) – shape = (batchsize, num_classes).
Binary mask indicating whether a class is present in the image patches.
mask_class_not_present[k]==1means that the image does not contain examples of classk. Then for all pixels in the patch, classkpredictive probabilities are trained towards0. Limit case:mask_class_not_present[:]==0will result in the ordinary NLL. - name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
Examples
- A cube contains no class
k. Instead of labelling the remaining classes they can be marked as unlabelled by the first mask (mask_class_labeled[:]==0, whethermask_class_labeled[k]is0or1is actually indifferent because the labels should not bey==kanyway in this case). Additionallymask_class_not_present[k]==1(otherwise0) to suppress predictions ofkin in this patch. The actual value of the labels is indifferent, it can either be-1or it could be the background class, if the background is marked as unlabelled (i.e. then those labels are ignored). - Only part of the cube is densely labelled. Set
mask_class_labeled[:]=1for all classes, but set the label values in the unlabelled part to-1to ignore this part. - Only a particular class
kis labelled in the cube. Either set all other label pixels to-1or the corresponding flags inmask_class_labeledfor the unlabelled classes.
Note
Using
-1labels or telling that a class is not labelled, is somewhat redundant and just supported for convenience.
-
class
elektronn2.neuromancer.loss.MalisNLL(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeMalis NLL node. (See https://github.com/TuragaLab/malis)
Parameters: - pred (Node) – Prediction node.
- aff_gt (T.Tensor) –
- seg_gt (T.Tensor) –
- nhood (np.ndarray) –
- unrestrict_neg (bool) –
- class_weights (T.Tensor) – weight vector of float32 of length
n_lab. Values:1.0(default),w < 1.0(less important),w > 1.0(more important class). - example_weights (T.Tensor) – weight vector of float32 of shape
(bs, z, x, y)that can give the individual examples (i.e. labels for output pixels) different weights. Values:1.0(default),w < 1.0(less important),w > 1.0(more important example). Note: if this is not normalised/bounded it may result in a effectively modified learning rate! - name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
-
elektronn2.neuromancer.loss.Errors(pred, target, target_is_sparse=False, n_class='auto', n_indep='auto', name='errors', print_repr=True)[source]¶
-
class
elektronn2.neuromancer.loss.BetaNLL(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeSimilar to BinaryNLL loss but “modulated” in scale by the variance.
Parameters: Computes element-wise:
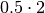
-
elektronn2.neuromancer.loss.SobelizedLoss(pred, target, loss_type='abs', loss_kwargs=None)[source]¶ SobelizedLoss node.
Parameters: - pred (Node) – Prediction node.
- target (T.Tensor) – corresponds to a vector that gives the correct label for each example. Labels < 0 are ignored (e.g. can be used for label propagation).
- loss_type (str) – Only “abs” is supported.
- loss_kwargs (dict) – kwargs for the AbsLoss constructor.
Returns: The loss node.
Return type:
-
class
elektronn2.neuromancer.loss.BlockedMultinoulliNLL(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeReturns the symbolic mean and instance-wise negative log-likelihood of the prediction of this model under a given target distribution.
Parameters: - pred (Node) – Prediction node.
- target (T.Tensor) – corresponds to a vector that gives the correct label for each example. Labels < 0 are ignored (e.g. can be used for label propagation).
- blocking_factor (float) – Blocking factor.
- target_is_sparse (bool) – If the target is sparse.
- class_weights (T.Tensor) – weight vector of float32 of length
n_lab. Values:1.0(default),w < 1.0(less important),w > 1.0(more important class). - example_weights (T.Tensor) – weight vector of float32 of shape
(bs, z, x, y)that can give the individual examples (i.e. labels for output pixels) different weights. Values:1.0(default),w < 1.0(less important),w > 1.0(more important example). Note: if this is not normalised/bounded it may result in a effectively modified learning rate! - following refers to lazy labels, the masks are always on a per patch basis, depending on the (The) –
- cube of the patch. The masks are properties of the individual image cubes and must be loaded (origin) –
- CNNData. (into) –
- mask_class_labeled (T.Tensor) – shape = (batchsize, num_classes).
Binary masks indicating whether a class is properly labeled in
y. If a classkis (in general) present in the image patches andmask_class_labeled[k]==1, then the labels must obeyy==kfor all pixels where the class is present. If a classkis present in the image, but was not labeled (-> cheaper labels), setmask_class_labeled[k]=0. Then all pixels for which they==kwill be ignored. Alternative: sety=-1to ignore those pixels. Limit case:mask_class_labeled[:]==1will result in the ordinary NLL. - mask_class_not_present (T.Tensor) – shape = (batchsize, num_classes).
Binary mask indicating whether a class is present in the image patches.
mask_class_not_present[k]==1means that the image does not contain examples of classk. Then for all pixels in the patch, classkpredictive probabilities are trained towards0. Limit case:mask_class_not_present[:]==0will result in the ordinary NLL. - name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
Examples
- A cube contains no class
k. Instead of labelling the remaining classes they can be marked as unlabelled by the first mask (mask_class_labeled[:]==0, whethermask_class_labeled[k]is0or1is actually indifferent because the labels should not bey==kanyway in this case). Additionallymask_class_not_present[k]==1(otherwise0) to suppress predictions ofkin in this patch. The actual value of the labels is indifferent, it can either be-1or it could be the background class, if the background is marked as unlabelled (i.e. then those labels are ignored). - Only part of the cube is densely labelled. Set
mask_class_labeled[:]=1for all classes, but set the label values in the unlabelled part to-1to ignore this part. - Only a particular class
kis labelled in the cube. Either set all other label pixels to-1or the corresponding flags inmask_class_labeledfor the unlabelled classes.
Note
Using
-1labels or telling that a class is not labelled, is somewhat redundant and just supported for convenience.
-
class
elektronn2.neuromancer.loss.OneHot(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeOnehot node.
Parameters: - target (T.Tensor) – Target tensor.
- n_class (int) –
- axis –
- name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
-
class
elektronn2.neuromancer.loss.EuclideanDistance(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeEuclidean distance node.
Parameters: - pred (Node) – Prediction node.
- target (T.Tensor) – corresponds to a vector that gives the correct label for each example. Labels < 0 are ignored (e.g. can be used for label propagation).
- margin (float/None) –
- scale_correction (float/None) – Downweights absolute deviations for large target scale. The value specifies the target value at which the square deviation has half weight compared to target=0 If the target is twice as large as this value the downweight is 1/3 and so on. Note: the smaller this value the stronger the effect. No effect would be +inf
- name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.
-
class
elektronn2.neuromancer.loss.RampLoss(**kwargs)[source]¶ Bases:
elektronn2.neuromancer.node_basic.NodeRampLoss node.
Parameters: - pred (Node) – Prediction node.
- target (T.Tensor) – corresponds to a vector that gives the correct label for each example. Labels < 0 are ignored (e.g. can be used for label propagation).
- margin (float/None) –
- scale_correction (float/None) – downweights absolute deviations for large target scale. The value specifies the target value at which the square deviation has half weight compared to target=0 If the target is twice as large as this value the downweight is 1/3 and so on. Note: the smaller this value the stronger the effect. No effect would be +inf
- name (str) – Node name.
- print_repr (bool) – Whether to print the node representation upon initialisation.